
|
UMBC High Performance Computing Facility

How to run R programs on tara
[araim1@tara-fe1 ~]$ R --version R version 2.13.0 (2011-04-13) ... [araim1@tara-fe1 ~]$
[araim1@tara-fe1 ~]$ module load R [araim1@tara-fe1 ~]$ R --version R version 2.14.0 (2011-10-31) ... [araim1@tara-fe1 ~]$
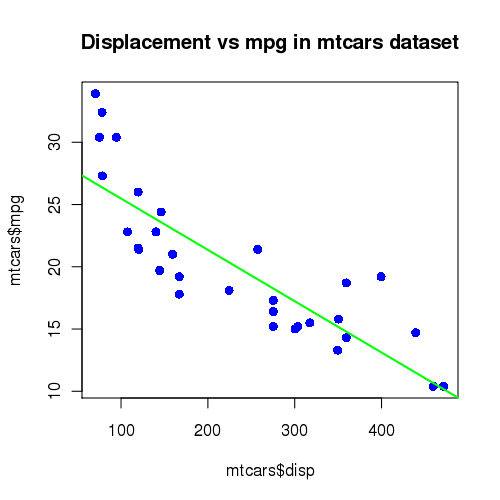
#!/bin/bash #SBATCH --job-name=plotR #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop Xvfb & export DISPLAY=:0 Rscript driver.R
[araim1@tara-fe1 plot-R]$ sbatch run.slurm sbatch: Submitted batch job 2623 [araim1@tara-fe1 plot-R]$
[araim1@tara-fe1 plot-R]$ ls mtcars.png slurm.out run.slurm slurm.err [araim1@tara-fe1 plot-R]$
[araim1@tara-fe1 ~]$ switcher mpi = gcc-openmpi-1.3.3-p1 [araim1@tara-fe1 ~]$ switcher_reload
R_LIBS=/usr/cluster/contrib/gcc-openmpi-1.3.3/Rlibs
R_LIBS=/usr/cluster/contrib/gcc-openmpi-1.3.3/Rlibs:/path/to/more/libs
[araim1@tara-fe1 ~]$ R R version 2.11.1 (2010-05-31) Copyright (C) 2010 The R Foundation for Statistical Computing ISBN 3-900051-07-0 ... > .libPaths() [1] "/usr/cluster/contrib/gcc-openmpi-1.3.3/Rlibs" [2] "/usr/lib64/R/library" > library(snow) > library(Rmpi)
library(snowslurm)
# Initialize SNOW using SOCKET communication, customized for SLURM
# Everything else is standard SNOW
cluster <- makeSLURMcluster()
# Print the hostname for each cluster member
sayhello <- function()
{
info <- Sys.info()[c("nodename", "machine")]
paste("Hello from", info[1], "with CPU type", info[2])
}
names <- clusterCall(cluster, sayhello)
print(unlist(names))
# Compute row sums in parallel using all processes,
# then a grand sum at the end on the master process
parallelSum <- function(m, n)
{
A <- matrix(rnorm(m*n), nrow = m, ncol = n)
row.sums <- parApply(cluster, A, 1, sum)
print(sum(row.sums))
}
parallelSum(500, 500)
stopCluster(cluster)
#!/bin/bash #SBATCH --job-name=snow-sock-test #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop #SBATCH --nodes=2 #SBATCH --ntasks-per-node=3 R --no-save < snow-test.R
[araim1@tara-fe1 R-snow-sock-test2]$ sbatch run.slurm sbatch: Submitted batch job 2648 [araim1@tara-fe1 R-snow-sock-test2]$
R version 3.0.2 (2013-09-25) -- "Frisbee Sailing"
Copyright (C) 2013 The R Foundation for Statistical Computing
Platform: x86_64-unknown-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(snowslurm)
>
> # Initialize SNOW using SOCKET communication, customized for SLURM
> # Everything else is standard SNOW
> cluster <- makeSLURMcluster()
2011-08-04 05:26:25 - About to spawn 6 remote slaves for SLURM SOCKET cluster
2011-08-04 05:26:26 - Remote cluster is constructed
>
> # Print the hostname for each cluster member
> sayhello <- function()
+ {
+ info <- Sys.info()[c("nodename", "machine")]
+ paste("Hello from", info[1], "with CPU type", info[2])
+ }
>
> names <- clusterCall(cluster, sayhello)
> print(unlist(names))
[1] "Hello from n1 with CPU type x86_64" "Hello from n1 with CPU type x86_64"
[3] "Hello from n1 with CPU type x86_64" "Hello from n2 with CPU type x86_64"
[5] "Hello from n2 with CPU type x86_64" "Hello from n2 with CPU type x86_64"
>
> # Compute row sums in parallel using all processes,
> # then a grand sum at the end on the master process
> parallelSum <- function(m, n)
+ {
+ A <- matrix(rnorm(m*n), nrow = m, ncol = n)
+ row.sums <- parApply(cluster, A, 1, sum)
+ print(sum(row.sums))
+ }
>
> parallelSum(500, 500)
[1] -341.2832
>
> stopCluster(cluster)
>
library(Rmpi)
library(snow)
# Initialize SNOW using MPI communication. The first line will get the
# number of MPI processes the scheduler assigned to us. Everything else
# is standard SNOW
np <- mpi.universe.size()
cluster <- makeMPIcluster(np)
# Print the hostname for each cluster member
sayhello <- function()
{
info <- Sys.info()[c("nodename", "machine")]
paste("Hello from", info[1], "with CPU type", info[2])
}
names <- clusterCall(cluster, sayhello)
print(unlist(names))
# Compute row sums in parallel using all processes,
# then a grand sum at the end on the master process
parallelSum <- function(m, n)
{
A <- matrix(rnorm(m*n), nrow = m, ncol = n)
row.sums <- parApply(cluster, A, 1, sum)
print(sum(row.sums))
}
parallelSum(500, 500)
stopCluster(cluster)
mpi.exit()
#!/bin/bash #SBATCH --job-name=snow-mpi-test #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop #SBATCH --nodes=2 #SBATCH --ntasks-per-node=3 mpirun -np 1 R --no-save < snow-test.R
[araim1@tara-fe1 R-snow-mpi-test]$ sbatch run.slurm sbatch: Submitted batch job 2648 [araim1@tara-fe1 R-snow-mpi-test]$
R version 3.0.2 (2013-09-25) -- "Frisbee Sailing"
Copyright (C) 2013 The R Foundation for Statistical Computing
Platform: x86_64-unknown-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(Rmpi)
> library(snow)
>
> # Initialize SNOW using MPI communication. The first line will get the
> # number of MPI processes the scheduler assigned to us. Everything else
> # is standard SNOW
>
> np <- mpi.universe.size()
> cluster <- makeMPIcluster(np)
6 slaves are spawned successfully. 0 failed.
>
> # Print the hostname for each cluster member
> sayhello <- function()
+ {
+ info <- Sys.info()[c("nodename", "machine")]
+ paste("Hello from", info[1], "with CPU type", info[2])
+ }
>
> names <- clusterCall(cluster, sayhello)
> print(unlist(names))
[1] "Hello from n1 with CPU type x86_64" "Hello from n1 with CPU type x86_64"
[3] "Hello from n2 with CPU type x86_64" "Hello from n2 with CPU type x86_64"
[5] "Hello from n2 with CPU type x86_64" "Hello from n1 with CPU type x86_64"
>
> # Compute row sums in parallel using all processes,
> # then a grand sum at the end on the master process
> parallelSum <- function(m, n)
+ {
+ A <- matrix(rnorm(m*n), nrow = m, ncol = n)
+ row.sums <- parApply(cluster, A, 1, sum)
+ print(sum(row.sums))
+ }
>
> parallelSum(500, 500)
[1] 166.6455
>
> stopCluster(cluster)
[1] 1
> mpi.exit()
[1] "Detaching Rmpi. Rmpi cannot be used unless relaunching R."
>
>
In the current section, we show how to run Rmpi in the master/slave mode. In the next section, we switch to the single program multiple data paradigm. We have noted that the performance of master/slave mode on tara is significantly worse than SPMD mode, but the reason is not yet apparent.
Rmpi features many familiar communications like "bcast" and "send". For more information about using Rmpi, a good place to check is the reference manual on CRAN. Also see this tutorial.library(Rmpi)
mpi.spawn.Rslaves(needlog = FALSE)
mpi.bcast.cmd( id <- mpi.comm.rank() )
mpi.bcast.cmd( np <- mpi.comm.size() )
mpi.bcast.cmd( host <- mpi.get.processor.name() )
result <- mpi.remote.exec(paste("I am", id, "of", np, "running on", host))
print(unlist(result))
mpi.close.Rslaves(dellog = FALSE)
mpi.exit()
An MPI process has executed an operation involving a call to the "fork()" system call to create a child process. Open MPI is currently operating in a condition that could result in memory corruption or other system errors...The option "needlog = FALSE" tells Rmpi not to create the extra log files associated with this cleanup. If you would like to see the log files along with your results, do not specify this option.
#!/bin/bash #SBATCH --job-name=Rmpi_hello #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop #SBATCH --nodes=2 #SBATCH --ntasks-per-node=4 mpirun -np 1 R --no-save < hello.R
[araim1@maya-usr1 Rmpi-test]$ cat slurm.out
[... R disclaimer message ...]
> library(Rmpi)
> mpi.spawn.Rslaves(needlog = FALSE)
8 slaves are spawned successfully. 0 failed.
master (rank 0, comm 1) of size 9 is running on: n1
slave1 (rank 1, comm 1) of size 9 is running on: n1
slave2 (rank 2, comm 1) of size 9 is running on: n1
slave3 (rank 3, comm 1) of size 9 is running on: n1
slave4 (rank 4, comm 1) of size 9 is running on: n2
slave5 (rank 5, comm 1) of size 9 is running on: n2
slave6 (rank 6, comm 1) of size 9 is running on: n2
slave7 (rank 7, comm 1) of size 9 is running on: n2
slave8 (rank 8, comm 1) of size 9 is running on: n1
>
> mpi.bcast.cmd( id <- mpi.comm.rank() )
> mpi.bcast.cmd( np <- mpi.comm.size() )
> mpi.bcast.cmd( host <- mpi.get.processor.name() )
> result <- mpi.remote.exec(paste("I am", id, "of", np, "running on", host))
>
> print(unlist(result))
slave1 slave2
"I am 1 of 9 running on n1" "I am 2 of 9 running on n1"
slave3 slave4
"I am 3 of 9 running on n1" "I am 4 of 9 running on n2"
slave5 slave6
"I am 5 of 9 running on n2" "I am 6 of 9 running on n2"
slave7 slave8
"I am 7 of 9 running on n2" "I am 8 of 9 running on n1"
>
> mpi.close.Rslaves(dellog = FALSE)
[1] 1
> mpi.exit()
[1] "Detaching Rmpi. Rmpi cannot be used unless relaunching R."
>
library(Rmpi)
mpi.spawn.Rslaves(needlog = FALSE)
mpi.bcast.cmd( id <- mpi.comm.rank() )
mpi.bcast.cmd( np <- mpi.comm.size() )
mpi.bcast.cmd( host <- mpi.get.processor.name() )
result <- mpi.remote.exec(paste("I am", id, "of", np, "running on", host))
print(unlist(result))
# Sample one normal observation on the master and each slave
x <- rnorm(1)
mpi.bcast.cmd(x <- rnorm(1))
# Gather the entire x vector (by default to process 0, the master)
mpi.bcast.cmd(mpi.gather.Robj(x))
y <- mpi.gather.Robj(x)
print(unlist(y))
# Sum the x vector together, storing the result on process 0 by default
mpi.bcast.cmd(mpi.reduce(x, op = "sum"))
z <- mpi.reduce(x, op = "sum")
print(z)
mpi.close.Rslaves(dellog = FALSE)
mpi.exit()
#!/bin/bash #SBATCH --job-name=Rmpi_test #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop #SBATCH --nodes=1 #SBATCH --ntasks-per-node=8 mpirun -np 1 R --no-save < driver.R
[araim1@maya-usr1 Rmpi-comm-test]$ cat slurm.out
[... R disclaimer message ...]
> library(Rmpi)
>
> mpi.spawn.Rslaves(needlog = FALSE)
8 slaves are spawned successfully. 0 failed.
master (rank 0, comm 1) of size 9 is running on: n1
slave1 (rank 1, comm 1) of size 9 is running on: n1
... ... ...
slave8 (rank 8, comm 1) of size 9 is running on: n1
>
> mpi.bcast.cmd( id <- mpi.comm.rank() )
> mpi.bcast.cmd( np <- mpi.comm.size() )
> mpi.bcast.cmd( host <- mpi.get.processor.name() )
> result <- mpi.remote.exec(paste("I am", id, "of", np, "running on", host))
> print(unlist(result))
slave1 slave2
"I am 1 of 9 running on n1" "I am 2 of 9 running on n1"
slave3 slave4
"I am 3 of 9 running on n1" "I am 4 of 9 running on n1"
slave5 slave6
"I am 5 of 9 running on n1" "I am 6 of 9 running on n1"
slave7 slave8
"I am 7 of 9 running on n1" "I am 8 of 9 running on n1"
>
> # Sample one normal observation on the master and each slave
> x <- rnorm(1)
> mpi.bcast.cmd(x <- rnorm(1))
>
> # Gather the entire x vector (by default to process 0, the master)
> mpi.bcast.cmd(mpi.gather.Robj(x))
> y <- mpi.gather.Robj(x)
> print(unlist(y))
[1] -2.6498050 0.5241441 -0.6747354 0.5915066 0.7660781 0.3608518 -2.7048508
[8] -0.4686277 0.5241441
>
> # Sum the x vector together, storing the result on process 0 by default
> mpi.bcast.cmd(mpi.reduce(x, op = "sum"))
> z <- mpi.reduce(x, op = "sum")
> print(z)
[1] -3.731294
>
> mpi.close.Rslaves(dellog = FALSE)
[1] 1
> mpi.exit()
[1] "Detaching Rmpi. Rmpi cannot be used unless relaunching R."
>
library(Rmpi)
id <- mpi.comm.rank(comm = 0)
np <- mpi.comm.size(comm = 0)
hostname <- mpi.get.processor.name()
msg <- sprintf("Hello world from process %03d of %03d, on host %s\n",
id, np, hostname)
cat(msg)
mpi.barrier(comm = 0)
mpi.finalize()
#!/bin/bash #SBATCH --job-name=R_MPI #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop #SBATCH --nodes=2 #SBATCH --ntasks-per-node=4 mpirun Rscript hello.R
[araim1@maya-usr1 Rmpi-hello-spmd]$ sbatch run.slurm [araim1@maya-usr1 Rmpi-hello-spmd]$ cat slurm.err [araim1@maya-usr1 Rmpi-hello-spmd]$ cat slurm.out Hello world from process 001 of 016, on host n1 Hello world from process 003 of 016, on host n1 Hello world from process 004 of 016, on host n1 Hello world from process 010 of 016, on host n2 Hello world from process 011 of 016, on host n2 Hello world from process 013 of 016, on host n2 Hello world from process 009 of 016, on host n2 Hello world from process 002 of 016, on host n1 Hello world from process 008 of 016, on host n2 Hello world from process 012 of 016, on host n2 Hello world from process 014 of 016, on host n2 Hello world from process 000 of 016, on host n1 Hello world from process 015 of 016, on host n2 Hello world from process 007 of 016, on host n1 [araim1@maya-usr1 Rmpi-hello-spmd]$
library(Rmpi)
id <- mpi.comm.rank(comm = 0)
np <- mpi.comm.size(comm = 0)
hostname <- mpi.get.processor.name()
if (id == 0) {
msg <- sprintf("Hello world from process %03d", id)
cat("Process 0: Received msg from process 0 saying:", msg, "\n")
for (i in seq(1, np-1)) {
# buf is just a buffer to receive a string
buf <- paste(rep(" ", 64), collapse="")
recv <- mpi.recv(x = buf, type = 3, source = i, tag = 0, comm = 0)
cat("Process 0: Received msg from process", i, "saying:", recv, "\n")
}
} else {
msg <- sprintf("Hello world from process %03d", id)
mpi.send(msg, 3, dest = 0, tag = 0, comm = 0)
}
mpi.barrier(comm = 0)
mpi.finalize()
#!/bin/bash #SBATCH --job-name=R_MPI #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop #SBATCH --nodes=2 #SBATCH --ntasks-per-node=8 mpirun Rscript driver.R
[araim1@maya-usr1 Rmpi-hellosendrecv-spmd]$ sbatch run.slurm [araim1@maya-usr1 Rmpi-hellosendrecv-spmd]$ cat slurm.err [araim1@maya-usr1 Rmpi-hellosendrecv-spmd]$ cat slurm.out Process 0: Received msg from process 0 saying: Hello world from process 000 Process 0: Received msg from process 1 saying: Hello world from process 001 Process 0: Received msg from process 2 saying: Hello world from process 002 Process 0: Received msg from process 3 saying: Hello world from process 003 Process 0: Received msg from process 4 saying: Hello world from process 004 Process 0: Received msg from process 5 saying: Hello world from process 005 Process 0: Received msg from process 6 saying: Hello world from process 006 Process 0: Received msg from process 7 saying: Hello world from process 007 Process 0: Received msg from process 8 saying: Hello world from process 008 Process 0: Received msg from process 9 saying: Hello world from process 009 Process 0: Received msg from process 10 saying: Hello world from process 010 Process 0: Received msg from process 11 saying: Hello world from process 011 Process 0: Received msg from process 12 saying: Hello world from process 012 Process 0: Received msg from process 13 saying: Hello world from process 013 Process 0: Received msg from process 14 saying: Hello world from process 014 Process 0: Received msg from process 15 saying: Hello world from process 015 [araim1@maya-usr1 Rmpi-hellosendrecv-spmd]$
library(Rmpi)
id <- mpi.comm.rank(comm = 0)
np <- mpi.comm.size(comm = 0)
hostname <- mpi.get.processor.name()
# Generate k normal observations per process
k <- 5
x <- rnorm(k)
# Combine the x vectors together on every process
gather.result <- mpi.allgather(x, type = 2,
rdata = numeric(k * np), comm = 0)
# Log out for each process:
# 1) The local part x
# 2) The combined vector gather.result
logfile <- sprintf("process-%03d.log", id)
sink(logfile)
cat("local x:\n")
print(x)
cat("\ngather.result:\n")
print(gather.result)
sink(NULL)
mpi.barrier(comm = 0)
mpi.finalize()
#!/bin/bash #SBATCH --job-name=R_MPI #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop #SBATCH --nodes=1 #SBATCH --ntasks-per-node=3 mpirun Rscript driver.R
[araim1@maya-usr1 Rmpi-allgather-spmd]$ sbatch run.slurm [araim1@maya-usr1 Rmpi-allgather-spmd]$ cat slurm.err [araim1@maya-usr1 Rmpi-allgather-spmd]$ cat slurm.out [1] 1 [1] 1 [1] 1 [1][1] "Exiting Rmpi. Rmpi cannot be used unless relaunching R." "Exiting Rmpi. Rmpi cannot be used unless relaunching R." [1] "Exiting Rmpi. Rmpi cannot be used unless relaunching R." [1] 1 [1] 1 [1] 1 [araim1@maya-usr1 all-gather]$ cat process-000.log local x: [1] 0.5997779 0.6245255 -0.8847268 -3.1556043 -0.5805308 gather.result: [1] 0.59977794 0.62452554 -0.88472684 -3.15560425 -0.58053082 -0.82290441 [7] -0.75349182 -0.08821349 -0.97613294 0.88190176 0.59724681 -1.91898345 [13] -0.09221058 -0.11054082 0.85566243 [araim1@maya-usr1 all-gather]$ cat process-001.log local x: [1] -0.82290441 -0.75349182 -0.08821349 -0.97613294 0.88190176 gather.result: [1] 0.59977794 0.62452554 -0.88472684 -3.15560425 -0.58053082 -0.82290441 [7] -0.75349182 -0.08821349 -0.97613294 0.88190176 0.59724681 -1.91898345 [13] -0.09221058 -0.11054082 0.85566243 [araim1@maya-usr1 all-gather]$ cat process-002.log local x: [1] 0.59724681 -1.91898345 -0.09221058 -0.11054082 0.85566243 gather.result: [1] 0.59977794 0.62452554 -0.88472684 -3.15560425 -0.58053082 -0.82290441 [7] -0.75349182 -0.08821349 -0.97613294 0.88190176 0.59724681 -1.91898345 [13] -0.09221058 -0.11054082 0.8556624 [araim1@maya-usr1 Rmpi-allgather-spmd]$
library(pbdMPI, quiet = TRUE)
init()
.comm.size <- comm.size()
.comm.rank <- comm.rank()
msg <- sprintf("Hello world from process %d\n", .comm.rank)
comm.cat("Say hello:\n", quiet = TRUE)
comm.cat(msg, all.rank = TRUE)
k <- 10
x <- rep(.comm.rank, k)
comm.cat("\nOriginal x vector:\n", quiet = TRUE)
comm.print(x, all.rank = TRUE)
y <- allgather(x, unlist = TRUE)
A <- matrix(y, nrow = k, byrow = FALSE)
comm.cat("\nAllgather matrix (only showing process 0):\n", quiet = TRUE)
comm.print(A)
finalize()
#!/bin/bash #SBATCH --job-name=pbdMPI #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop #SBATCH --nodes=2 #SBATCH --ntasks-per-node=3 mpirun Rscript driver.R
[araim1@maya-usr1 helloMPI]$ sbatch run.slurm
[araim1@maya-usr1 helloMPI]$ cat slurm.err
[araim1@maya-usr1 helloMPI]$ cat slurm.out
Say hello:
COMM.RANK = 0
Hello world from process 0
COMM.RANK = 1
Hello world from process 1
COMM.RANK = 2
Hello world from process 2
COMM.RANK = 3
Hello world from process 3
COMM.RANK = 4
Hello world from process 4
COMM.RANK = 5
Hello world from process 5
Original x vector:
COMM.RANK = 0
[1] 0 0 0 0 0 0 0 0 0 0
COMM.RANK = 1
[1] 1 1 1 1 1 1 1 1 1 1
COMM.RANK = 2
[1] 2 2 2 2 2 2 2 2 2 2
COMM.RANK = 3
[1] 3 3 3 3 3 3 3 3 3 3
COMM.RANK = 4
[1] 4 4 4 4 4 4 4 4 4 4
COMM.RANK = 5
[1] 5 5 5 5 5 5 5 5 5 5
Allgather matrix (only showing process 0):
COMM.RANK = 0
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 0 1 2 3 4 5
[2,] 0 1 2 3 4 5
[3,] 0 1 2 3 4 5
[4,] 0 1 2 3 4 5
[5,] 0 1 2 3 4 5
[6,] 0 1 2 3 4 5
[7,] 0 1 2 3 4 5
[8,] 0 1 2 3 4 5
[9,] 0 1 2 3 4 5
[10,] 0 1 2 3 4 5
[araim1@maya-usr1 helloMPI]$
[araim1@tara-fe1 ~]$ R R version x.xx.x (20xx-xx-xx) ... > demo(package = "pbdMPI") > demo(package = "pbdDMAT") > demo(package = "pbdSLAP") > demo(package = "pbdDEMO")
Demos in package 'pbdMPI': allgather pbdMPI An example of allgather. allreduce pbdMPI An example of allreduce. any_all pbdMPI An example of any and all. bcast pbdMPI An example of bcast. divide pbdMPI An example of dividing jobs. gather pbdMPI An example of gather. pbdApply pbdMPI An example of parallel apply. pbdLapply pbdMPI An example of parallel lapply. reduce pbdMPI An example of reduce. scatter pbdMPI An example of scatter. seed pbdMPI An example of random number generation. sort pbdMPI An example of sorting.
#!/bin/bash #SBATCH --job-name=pbdDemo #SBATCH --output=slurm.out #SBATCH --error=slurm.err #SBATCH --partition=develop #SBATCH --nodes=1 #SBATCH --ntasks-per-node=3 mpirun Rscript -e "demo(scatter, 'pbdMPI', ask=F, echo=F)"